gmmcTrain
Train a GMM classifier
Contents
Syntax
- [cPrm, logLike] = gmmcTrain(DS)
- [cPrm, logLike] = gmmcTrain(DS, opt)
Description
[cPrm, logLike] = gmmcTrain(DS) returns the parameters of a GMM classifier based on the training of the give dataset DS.
- DS: Design dataset
- opt: GMMC options
- opt.arch.gaussianNum: A column vector indicating no. of Gaussians for each class
- opt.arch.covType: Type of covariance matrix
- tmmcOpt.train: Parameters for training each GMM, which can be obtained via gmmcTrain('defaultOpt').
- cPrm: Parameters for GMM classifier
- cPrm.gmm(i): Parameters for class i, which is modeled as a GMM
- cPrm.gmm(i).gmmPrm(j).mu: a mean vector of dim x 1 for Gaussian component j
- cPrm.gmm(i).gmmPrm(j).sigma: a covariance matrix for Gaussian component j
- cPrm.gmm(i).gmmPrm(j).w: a weighting factor for Gaussian component j
- cPrm.prior: Vector of priors, or simply the vector holding no. of entries in each class
- (To obtain the class sizes, you can use "dsClassSize".
- cPrm.gmm(i): Parameters for class i, which is modeled as a GMM
- logLike: Vector of log likelihood during training
Example
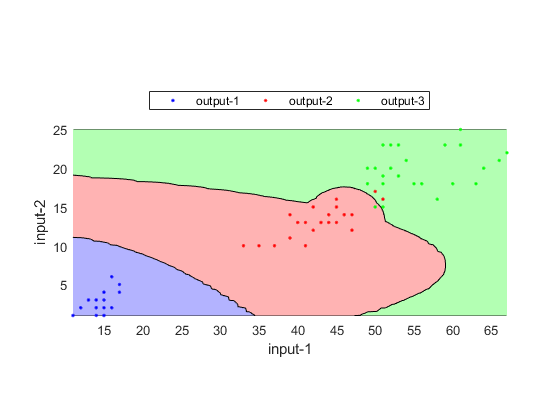
DS=prData('iris'); DS.input=DS.input(3:4, :); trainSet.input=DS.input(:, 1:2:end); trainSet.output=DS.output(:, 1:2:end); testSet.input=DS.input(:, 2:2:end); testSet.output=DS.output(:, 2:2:end); opt=gmmcTrain('defaultOpt'); cPrm=gmmcTrain(trainSet, opt); cOutput=gmmcEval(trainSet, cPrm); recogRate1=sum(trainSet.output==cOutput)/length(trainSet.output); fprintf('Inside-test recog. rate = %g%%\n', recogRate1*100); cOutput=gmmcEval(testSet, cPrm); recogRate2=sum(testSet.output==cOutput)/length(testSet.output); fprintf('Outside-test recog. rate = %g%%\n', recogRate2*100); TS.hitIndex=find(testSet.output==cOutput); gmmcPlot(testSet, cPrm, 'decBoundary');
Inside-test recog. rate = 97.3333% Outside-test recog. rate = 96%