pcaPerfViaKnncLoo
PCA analysis using KNNC and LOO
Contents
Syntax
- recogRate=pcaPerfViaKnncLoo(DS)
- recogRate=pcaPerfViaKnncLoo(DS, [], plotOpt)
- recogRate=pcaPerfViaKnncLoo(DS, maxDim, plotOpt)
Description
recogRate=pcaPerfViaKnncLoo(DS) return the leave-one-out recognition rate of KNNC on the dataset DS after dimension reduction using PCA (principal component analysis)
Example
PCA over WINE dataset
DS=prData('wine');
recogRate1=pcaPerfViaKnncLoo(DS, [], 1);
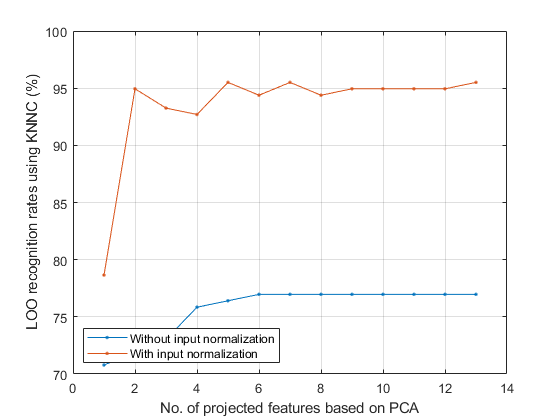
LOO recog. rate of KNNC using 1 dim = 126/178 = 70.7865% LOO recog. rate of KNNC using 2 dim = 128/178 = 71.9101% LOO recog. rate of KNNC using 3 dim = 130/178 = 73.0337% LOO recog. rate of KNNC using 4 dim = 135/178 = 75.8427% LOO recog. rate of KNNC using 5 dim = 136/178 = 76.4045% LOO recog. rate of KNNC using 6 dim = 137/178 = 76.9663% LOO recog. rate of KNNC using 7 dim = 137/178 = 76.9663% LOO recog. rate of KNNC using 8 dim = 137/178 = 76.9663% LOO recog. rate of KNNC using 9 dim = 137/178 = 76.9663% LOO recog. rate of KNNC using 10 dim = 137/178 = 76.9663% LOO recog. rate of KNNC using 11 dim = 137/178 = 76.9663% LOO recog. rate of KNNC using 12 dim = 137/178 = 76.9663% LOO recog. rate of KNNC using 13 dim = 137/178 = 76.9663%

Effect of input normalization of PCA over WINE dataset
DS=prData('wine'); recogRate1=pcaPerfViaKnncLoo(DS); DS.input=inputNormalize(DS.input); recogRate2=pcaPerfViaKnncLoo(DS); plot(1:length(recogRate1), 100*recogRate1, '.-', 1:length(recogRate2), 100*recogRate2, '.-'); grid on xlabel('No. of projected features based on PCA'); ylabel('LOO recognition rates using KNNC (%)'); legend('Without input normalization', 'With input normalization', 'location', 'southwest');