Tutorial on Fatty Liver Recognition (by Roger Jang)
Contents
This tutorial explains the basics of fatty liver recognition based on patients' data.
Preprocessing
Before we start, let's add necessary toolboxes to the search path of MATLAB:
addpath d:/users/jang/matlab/toolbox/utility addpath d:/users/jang/matlab/toolbox/sap addpath d:/users/jang/matlab/toolbox/machineLearning clear all; close all;
All the above toolboxes can be downloaded from the author's toolbox page. Make sure you are using the latest toolboxes to work with this script.
For compatibility, here we list the platform and MATLAB version that we used to run this script:
fprintf('Platform: %s\n', computer); fprintf('MATLAB version: %s\n', version); fprintf('Date & time: %s\n', char(datetime)); scriptStartTime=tic; % Timing for the whole script
Platform: PCWIN64 MATLAB version: 9.9.0.1467703 (R2020b) Date & time: 24-Apr-2022 23:27:48
Data preparation
First of all, we shall read the data from a data file:
file='D:\dataSet\mj\liver\small\fliver.csv'; fprintf('Reading %s...\n', file); tic; data=readtable(file); time=toc; fprintf('Time=%g sec\n', time); fprintf('Data size=%s\n', mat2str(size(data)));
Reading D:\dataSet\mj\liver\small\fliver.csv... Time=3.6498 sec Data size=[554202 35]
We delete some of the fields, as follows.
- Delete derived fields. See for "新創變項codebook.xlsx" details.
fieldToDelete={'bmi', 'ms1', 'ms2', 'ms3', 'ms4', 'ms5', 'ms'};
data=removevars(data, fieldToDelete);
fprintf('Data size=%s\n', mat2str(size(data)));
Data size=[554202 28]
- Delete apparently useless fields
fieldToDelete={'id', 'n', 'flivero', 'father', 'gfather_f', 'gfather_m', 'mother', 'gmother_f', 'gmother_m', 'marriage_98', 'marriage_14'};
data=removevars(data, fieldToDelete);
fprintf('Data size=%s\n', mat2str(size(data)));
Data size=[554202 17]
- Delete fields with too many NaN
fieldToDelete={'fincome', 'pincome'}; % Famile and personal income
data=removevars(data, fieldToDelete);
fprintf('Data size=%s\n', mat2str(size(data)));
Data size=[554202 15]
Since there are still numerous NaN in the dataset, we can eliminate data entries with NaN.
fprintf('List of no. of NaN at each field:\n'); fieldNames=fieldnames(data); feaNames=fieldNames(1:end-3); % The last 3 fields are not real. for i=1:length(feaNames) feaName=feaNames{i}; id=find(isnan(data.(feaName))); count=length(id); if count>0, fprintf('Nan count in data.%s=%d\n', feaName, count); end data(id,:)=[]; end fprintf('After removing data with fields of NaN, data size=%s\n', mat2str(size(data))); fprintf('Saving data.mat...\n'); save data data
List of no. of NaN at each field: Nan count in data.education=8670 Nan count in data.occupation=19668 Nan count in data.l_hdlc=16578 Nan count in data.systolic=1 Nan count in data.diastolic=10 After removing data with fields of NaN, data size=[509275 15] Saving data.mat...
Then we can create the basic dataset ds.
outputFieldName='fliver'; ds.output=data.(outputFieldName)'+1; % Starting from 1 data=removevars(data, outputFieldName); fieldNames=fieldnames(data); ds.inputName=fieldNames(1:end-3)'; [dataCount, feaCount]=size(data); ds.input=zeros(feaCount, dataCount); for i=1:feaCount ds.input(i,:)=data.(ds.inputName{i})'; end ds.outputName={'no', 'yes'}; disp(ds) fprintf('Saving ds.mat...\n'); save ds ds
output: [1×509275 double]
inputName: {1×14 cell}
input: [14×509275 double]
outputName: {'no' 'yes'}
Saving ds.mat...
We partition the whole dataset into training and test sets.
opt=cvDataGen('defaultOpt'); opt.foldNum=2; opt.cvDataType='full'; cvData=cvDataGen(ds, opt); dsTrain=cvData.TS; % Training set dsTest=cvData.VS; % Test set
For more detailed analysis, we partition the data based on gender.
opt=struct('inputName', 'gender', 'inputValue', 1); dsTrainFemale=dsSubset(dsTrain, opt); opt.inputValue=2; dsTrainMale=dsSubset(dsTrain, opt); opt.inputValue=1; dsTestFemale=dsSubset(dsTest, opt); opt.inputValue=2; dsTestMale=dsSubset(dsTest, opt);
We can plot the patient counts of the training data based on gender and years.
id=find(strcmp(dsTrainFemale.inputName, 'yr')); year=dsTrainFemale.input(id, :); figure; [a, b]=elementCount(year); subplot(211); bar(a, b); xlabel('Years'); ylabel('Patient counts'); title(sprintf('Female total=%d', size(dsTrainFemale.input,2))); year=dsTrainMale.input(id, :); [a, b]=elementCount(year); subplot(212); bar(a, b); xlabel('Years'); ylabel('Patient counts'); title(sprintf('Male total=%d', size(dsTrainMale.input,2))); axisLimitSame;
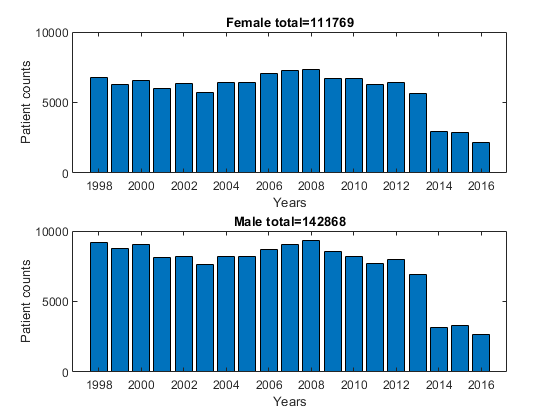
For simplicity, we shall use 2016 male data for further analysis.
opt=struct('inputName', 'yr', 'inputValue', 2016); ds=dsSubset(dsTrainMale, opt); % Training set ds dsSmallTest=dsSubset(dsTestMale, opt); % Test set dsSmallTest
ds =
struct with fields:
input: [12×2684 double]
output: [1×2684 double]
inputName: {1×12 cell}
outputName: {'no' 'yes'}
dsSmallTest =
struct with fields:
input: [12×2559 double]
output: [1×2559 double]
inputName: {1×12 cell}
outputName: {'no' 'yes'}
Dataset visualization
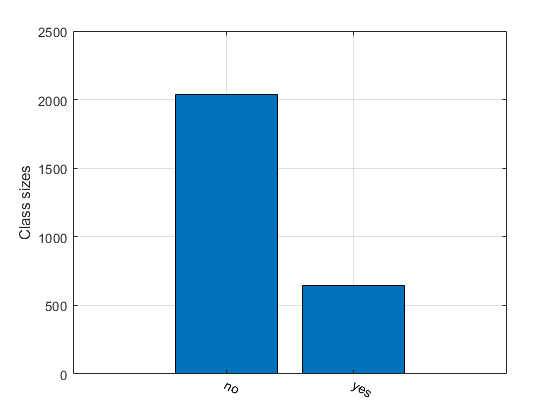
Size of the classes:
figure; [classSize, classLabel]=dsClassSize(ds, 1)
12 features
2684 instances
2 classes
classSize =
2039 645
classLabel =
1 2
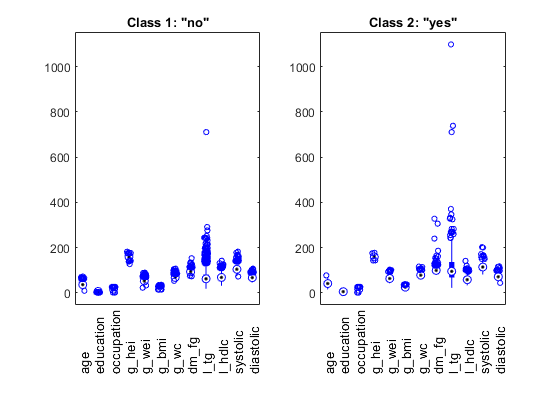
Box plot for two classes:
figure; dsBoxPlot(ds);
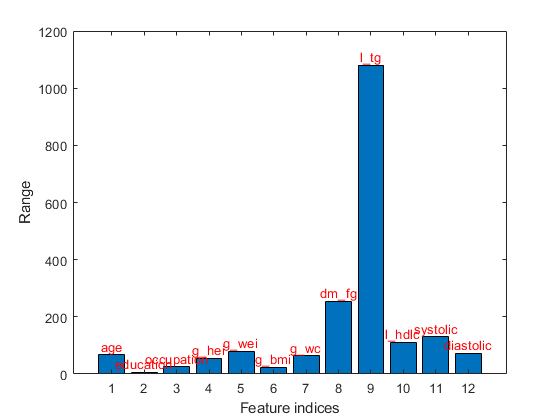
Range plot of the dataset:
figure; dsRangePlot(ds);
Range plot of the normalized dataset ds2:
ds2=ds; ds2.input=inputNormalize(ds2.input); figure; dsRangePlot(ds2);

Scatter plots of ds2:
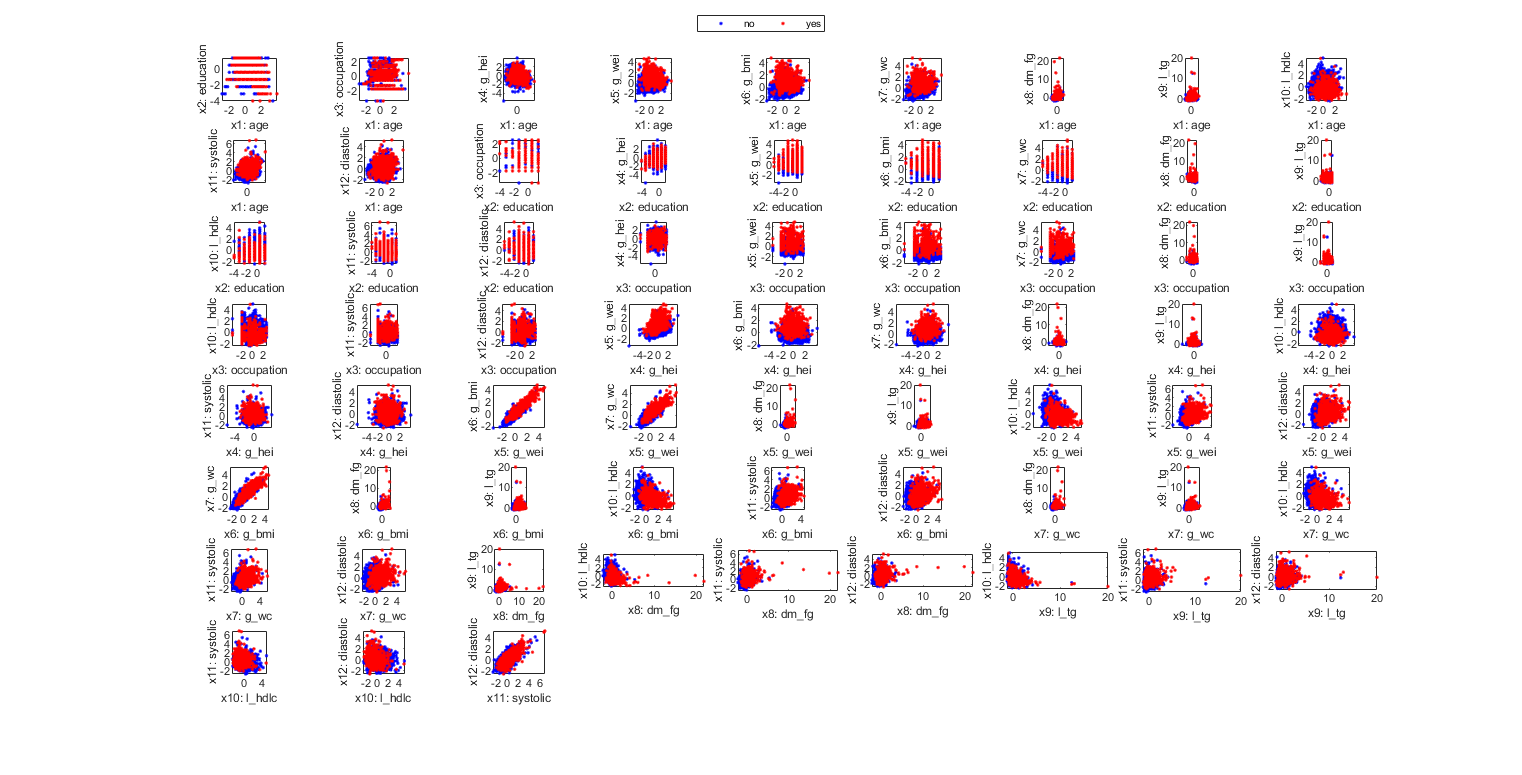
figure; dsProjPlot2(ds2); figEnlarge;
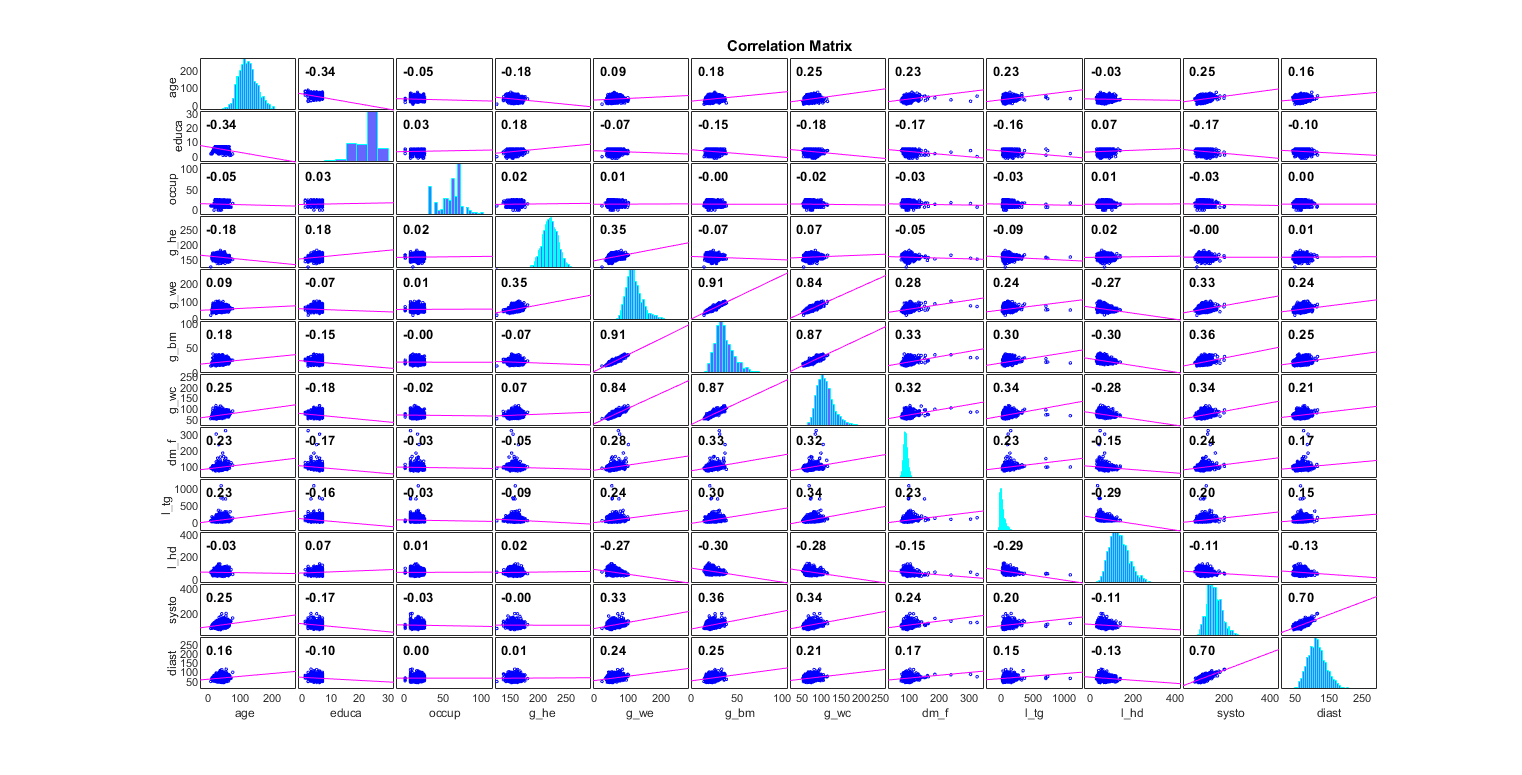
Correlation plot:
figure; corrplot(ds.input', 'varNames', strPurify(ds.inputName)); figEnlarge
Classification
KNN results on ds and ds2:
tic; rr=knncLoo(ds); fprintf('rr=%g%% for ds, time=%g sec\n', rr*100, toc); tic; rr=knncLoo(ds2); fprintf('rr=%g%% for ds2, time=%g sec\n', rr*100, toc);
rr=78.0551% for ds, time=0.205163 sec rr=78.465% for ds2, time=0.149496 sec
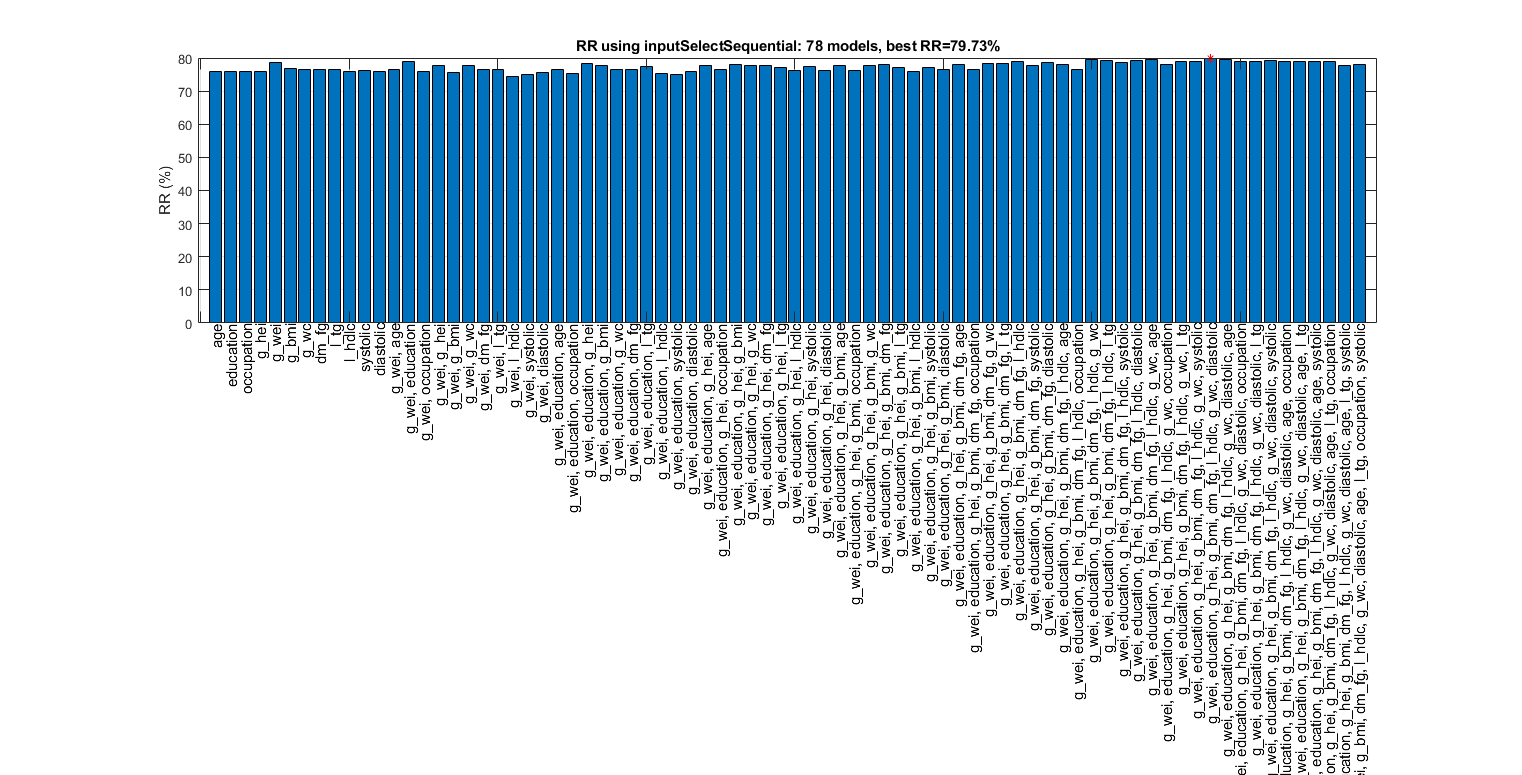
Sequential forward selection on the features of ds:
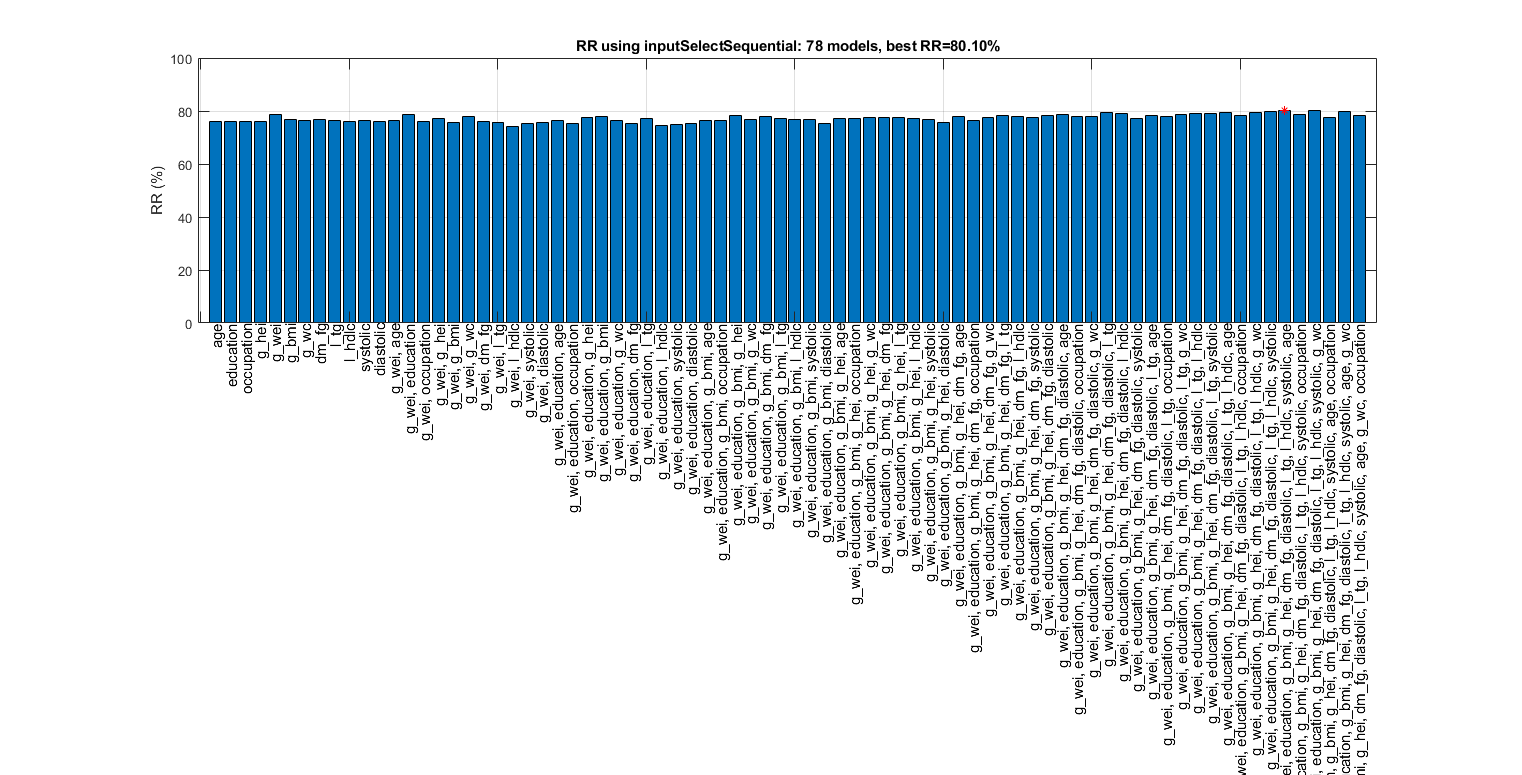
figure; tic; inputSelectSequential(ds, inf, 'knnc'); toc; figEnlarge
Construct 78 "knnc" models, each with up to 12 inputs selected from all 12 inputs...
Selecting input 1:
Model 1/78: selected={age} => Recog. rate = 75.89%
Model 2/78: selected={education} => Recog. rate = 75.93%
Model 3/78: selected={occupation} => Recog. rate = 75.97%
Model 4/78: selected={g_hei} => Recog. rate = 75.86%
Model 5/78: selected={g_wei} => Recog. rate = 78.69%
Model 6/78: selected={g_bmi} => Recog. rate = 76.83%
Model 7/78: selected={g_wc} => Recog. rate = 76.49%
Model 8/78: selected={dm_fg} => Recog. rate = 76.68%
Model 9/78: selected={l_tg} => Recog. rate = 76.49%
Model 10/78: selected={l_hdlc} => Recog. rate = 75.89%
Model 11/78: selected={systolic} => Recog. rate = 76.23%
Model 12/78: selected={diastolic} => Recog. rate = 76.04%
Currently selected inputs: g_wei => Recog. rate = 78.7%
Selecting input 2:
Model 13/78: selected={g_wei, age} => Recog. rate = 76.38%
Model 14/78: selected={g_wei, education} => Recog. rate = 78.80%
Model 15/78: selected={g_wei, occupation} => Recog. rate = 75.97%
Model 16/78: selected={g_wei, g_hei} => Recog. rate = 77.76%
Model 17/78: selected={g_wei, g_bmi} => Recog. rate = 75.75%
Model 18/78: selected={g_wei, g_wc} => Recog. rate = 77.83%
Model 19/78: selected={g_wei, dm_fg} => Recog. rate = 76.38%
Model 20/78: selected={g_wei, l_tg} => Recog. rate = 76.38%
Model 21/78: selected={g_wei, l_hdlc} => Recog. rate = 74.40%
Model 22/78: selected={g_wei, systolic} => Recog. rate = 74.89%
Model 23/78: selected={g_wei, diastolic} => Recog. rate = 75.75%
Currently selected inputs: g_wei, education => Recog. rate = 78.8%
Selecting input 3:
Model 24/78: selected={g_wei, education, age} => Recog. rate = 76.68%
Model 25/78: selected={g_wei, education, occupation} => Recog. rate = 75.30%
Model 26/78: selected={g_wei, education, g_hei} => Recog. rate = 78.32%
Model 27/78: selected={g_wei, education, g_bmi} => Recog. rate = 77.65%
Model 28/78: selected={g_wei, education, g_wc} => Recog. rate = 76.68%
Model 29/78: selected={g_wei, education, dm_fg} => Recog. rate = 76.64%
Model 30/78: selected={g_wei, education, l_tg} => Recog. rate = 77.42%
Model 31/78: selected={g_wei, education, l_hdlc} => Recog. rate = 75.22%
Model 32/78: selected={g_wei, education, systolic} => Recog. rate = 75.11%
Model 33/78: selected={g_wei, education, diastolic} => Recog. rate = 75.82%
Currently selected inputs: g_wei, education, g_hei => Recog. rate = 78.3%
Selecting input 4:
Model 34/78: selected={g_wei, education, g_hei, age} => Recog. rate = 77.79%
Model 35/78: selected={g_wei, education, g_hei, occupation} => Recog. rate = 76.42%
Model 36/78: selected={g_wei, education, g_hei, g_bmi} => Recog. rate = 78.09%
Model 37/78: selected={g_wei, education, g_hei, g_wc} => Recog. rate = 77.68%
Model 38/78: selected={g_wei, education, g_hei, dm_fg} => Recog. rate = 77.87%
Model 39/78: selected={g_wei, education, g_hei, l_tg} => Recog. rate = 77.05%
Model 40/78: selected={g_wei, education, g_hei, l_hdlc} => Recog. rate = 76.08%
Model 41/78: selected={g_wei, education, g_hei, systolic} => Recog. rate = 77.31%
Model 42/78: selected={g_wei, education, g_hei, diastolic} => Recog. rate = 76.34%
Currently selected inputs: g_wei, education, g_hei, g_bmi => Recog. rate = 78.1%
Selecting input 5:
Model 43/78: selected={g_wei, education, g_hei, g_bmi, age} => Recog. rate = 77.65%
Model 44/78: selected={g_wei, education, g_hei, g_bmi, occupation} => Recog. rate = 76.19%
Model 45/78: selected={g_wei, education, g_hei, g_bmi, g_wc} => Recog. rate = 77.79%
Model 46/78: selected={g_wei, education, g_hei, g_bmi, dm_fg} => Recog. rate = 78.02%
Model 47/78: selected={g_wei, education, g_hei, g_bmi, l_tg} => Recog. rate = 77.09%
Model 48/78: selected={g_wei, education, g_hei, g_bmi, l_hdlc} => Recog. rate = 75.82%
Model 49/78: selected={g_wei, education, g_hei, g_bmi, systolic} => Recog. rate = 77.16%
Model 50/78: selected={g_wei, education, g_hei, g_bmi, diastolic} => Recog. rate = 76.53%
Currently selected inputs: g_wei, education, g_hei, g_bmi, dm_fg => Recog. rate = 78.0%
Selecting input 6:
Model 51/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, age} => Recog. rate = 78.02%
Model 52/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, occupation} => Recog. rate = 76.60%
Model 53/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, g_wc} => Recog. rate = 78.43%
Model 54/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_tg} => Recog. rate = 78.46%
Model 55/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc} => Recog. rate = 78.95%
Model 56/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, systolic} => Recog. rate = 77.68%
Model 57/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, diastolic} => Recog. rate = 78.73%
Currently selected inputs: g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc => Recog. rate = 78.9%
Selecting input 7:
Model 58/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, age} => Recog. rate = 78.06%
Model 59/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, occupation} => Recog. rate = 76.60%
Model 60/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc} => Recog. rate = 79.69%
Model 61/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, l_tg} => Recog. rate = 79.10%
Model 62/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, systolic} => Recog. rate = 78.58%
Model 63/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, diastolic} => Recog. rate = 79.17%
Currently selected inputs: g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc => Recog. rate = 79.7%
Selecting input 8:
Model 64/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, age} => Recog. rate = 79.51%
Model 65/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, occupation} => Recog. rate = 78.17%
Model 66/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, l_tg} => Recog. rate = 78.95%
Model 67/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, systolic} => Recog. rate = 78.84%
Model 68/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic} => Recog. rate = 79.73%
Currently selected inputs: g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic => Recog. rate = 79.7%
Selecting input 9:
Model 69/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, age} => Recog. rate = 79.62%
Model 70/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, occupation} => Recog. rate = 78.84%
Model 71/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, l_tg} => Recog. rate = 78.87%
Model 72/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, systolic} => Recog. rate = 79.32%
Currently selected inputs: g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, age => Recog. rate = 79.6%
Selecting input 10:
Model 73/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, age, occupation} => Recog. rate = 78.84%
Model 74/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, age, l_tg} => Recog. rate = 78.91%
Model 75/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, age, systolic} => Recog. rate = 78.87%
Currently selected inputs: g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, age, l_tg => Recog. rate = 78.9%
Selecting input 11:
Model 76/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, age, l_tg, occupation} => Recog. rate = 78.99%
Model 77/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, age, l_tg, systolic} => Recog. rate = 77.87%
Currently selected inputs: g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, age, l_tg, occupation => Recog. rate = 79.0%
Selecting input 12:
Model 78/78: selected={g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, age, l_tg, occupation, systolic} => Recog. rate = 78.06%
Currently selected inputs: g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic, age, l_tg, occupation, systolic => Recog. rate = 78.1%
Overall maximal recognition rate = 79.7%.
Selected 8 inputs (out of 12): g_wei, education, g_hei, g_bmi, dm_fg, l_hdlc, g_wc, diastolic
Elapsed time is 7.329201 seconds.
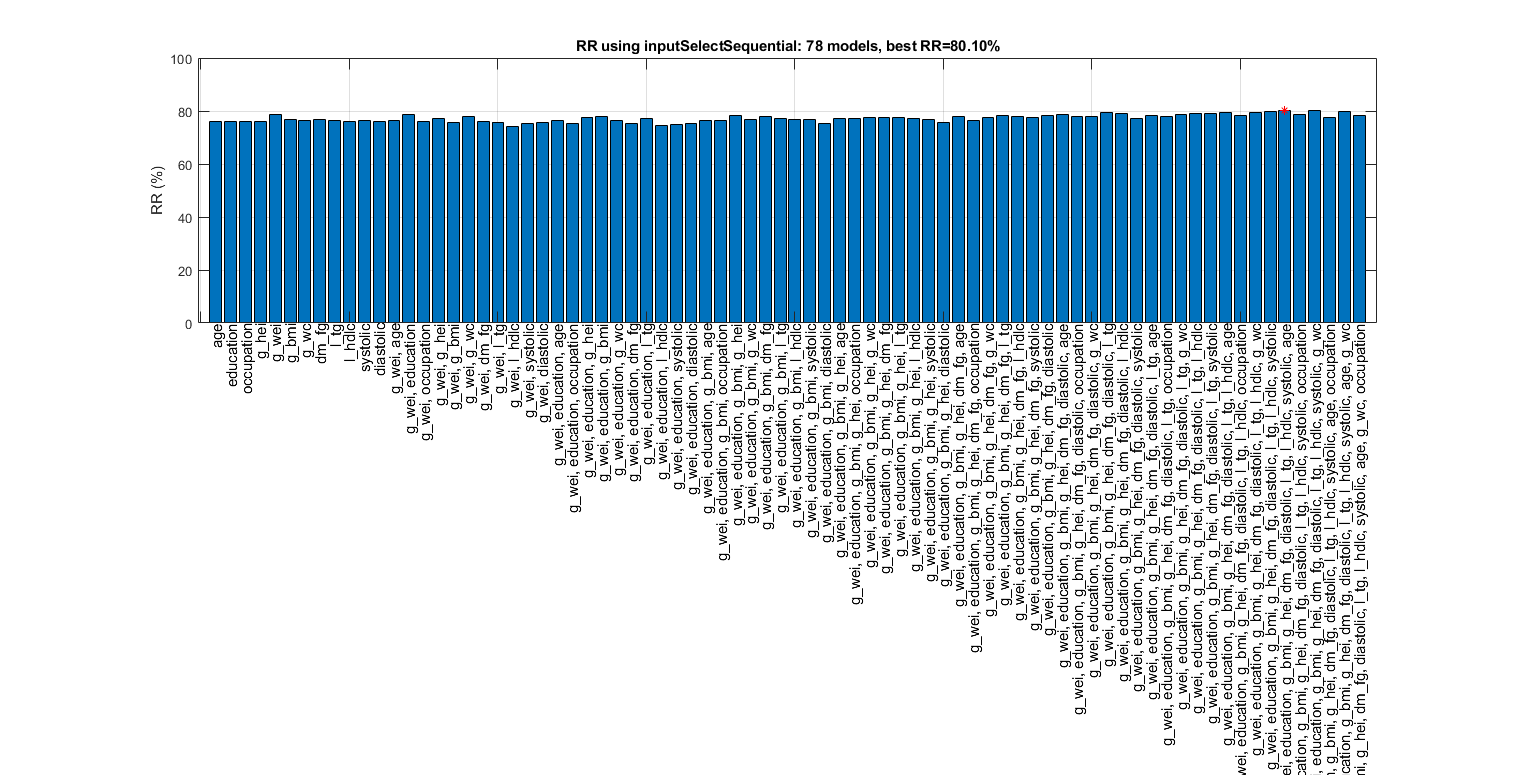
Sequential forward selection on the features of ds2:
figure; tic; inputSelectSequential(ds2, inf, 'knnc'); toc; figEnlarge
Construct 78 "knnc" models, each with up to 12 inputs selected from all 12 inputs...
Selecting input 1:
Model 1/78: selected={age} => Recog. rate = 75.89%
Model 2/78: selected={education} => Recog. rate = 75.93%
Model 3/78: selected={occupation} => Recog. rate = 75.97%
Model 4/78: selected={g_hei} => Recog. rate = 75.86%
Model 5/78: selected={g_wei} => Recog. rate = 78.69%
Model 6/78: selected={g_bmi} => Recog. rate = 76.83%
Model 7/78: selected={g_wc} => Recog. rate = 76.49%
Model 8/78: selected={dm_fg} => Recog. rate = 76.71%
Model 9/78: selected={l_tg} => Recog. rate = 76.56%
Model 10/78: selected={l_hdlc} => Recog. rate = 75.86%
Model 11/78: selected={systolic} => Recog. rate = 76.23%
Model 12/78: selected={diastolic} => Recog. rate = 75.97%
Currently selected inputs: g_wei => Recog. rate = 78.7%
Selecting input 2:
Model 13/78: selected={g_wei, age} => Recog. rate = 76.27%
Model 14/78: selected={g_wei, education} => Recog. rate = 78.73%
Model 15/78: selected={g_wei, occupation} => Recog. rate = 75.97%
Model 16/78: selected={g_wei, g_hei} => Recog. rate = 77.31%
Model 17/78: selected={g_wei, g_bmi} => Recog. rate = 75.78%
Model 18/78: selected={g_wei, g_wc} => Recog. rate = 77.79%
Model 19/78: selected={g_wei, dm_fg} => Recog. rate = 76.01%
Model 20/78: selected={g_wei, l_tg} => Recog. rate = 75.82%
Model 21/78: selected={g_wei, l_hdlc} => Recog. rate = 74.07%
Model 22/78: selected={g_wei, systolic} => Recog. rate = 75.34%
Model 23/78: selected={g_wei, diastolic} => Recog. rate = 75.67%
Currently selected inputs: g_wei, education => Recog. rate = 78.7%
Selecting input 3:
Model 24/78: selected={g_wei, education, age} => Recog. rate = 76.42%
Model 25/78: selected={g_wei, education, occupation} => Recog. rate = 75.34%
Model 26/78: selected={g_wei, education, g_hei} => Recog. rate = 77.72%
Model 27/78: selected={g_wei, education, g_bmi} => Recog. rate = 77.91%
Model 28/78: selected={g_wei, education, g_wc} => Recog. rate = 76.23%
Model 29/78: selected={g_wei, education, dm_fg} => Recog. rate = 75.41%
Model 30/78: selected={g_wei, education, l_tg} => Recog. rate = 77.31%
Model 31/78: selected={g_wei, education, l_hdlc} => Recog. rate = 74.48%
Model 32/78: selected={g_wei, education, systolic} => Recog. rate = 74.89%
Model 33/78: selected={g_wei, education, diastolic} => Recog. rate = 75.37%
Currently selected inputs: g_wei, education, g_bmi => Recog. rate = 77.9%
Selecting input 4:
Model 34/78: selected={g_wei, education, g_bmi, age} => Recog. rate = 76.60%
Model 35/78: selected={g_wei, education, g_bmi, occupation} => Recog. rate = 76.56%
Model 36/78: selected={g_wei, education, g_bmi, g_hei} => Recog. rate = 78.13%
Model 37/78: selected={g_wei, education, g_bmi, g_wc} => Recog. rate = 76.79%
Model 38/78: selected={g_wei, education, g_bmi, dm_fg} => Recog. rate = 77.94%
Model 39/78: selected={g_wei, education, g_bmi, l_tg} => Recog. rate = 77.12%
Model 40/78: selected={g_wei, education, g_bmi, l_hdlc} => Recog. rate = 76.97%
Model 41/78: selected={g_wei, education, g_bmi, systolic} => Recog. rate = 76.75%
Model 42/78: selected={g_wei, education, g_bmi, diastolic} => Recog. rate = 75.37%
Currently selected inputs: g_wei, education, g_bmi, g_hei => Recog. rate = 78.1%
Selecting input 5:
Model 43/78: selected={g_wei, education, g_bmi, g_hei, age} => Recog. rate = 77.01%
Model 44/78: selected={g_wei, education, g_bmi, g_hei, occupation} => Recog. rate = 77.09%
Model 45/78: selected={g_wei, education, g_bmi, g_hei, g_wc} => Recog. rate = 77.42%
Model 46/78: selected={g_wei, education, g_bmi, g_hei, dm_fg} => Recog. rate = 77.50%
Model 47/78: selected={g_wei, education, g_bmi, g_hei, l_tg} => Recog. rate = 77.42%
Model 48/78: selected={g_wei, education, g_bmi, g_hei, l_hdlc} => Recog. rate = 77.35%
Model 49/78: selected={g_wei, education, g_bmi, g_hei, systolic} => Recog. rate = 76.68%
Model 50/78: selected={g_wei, education, g_bmi, g_hei, diastolic} => Recog. rate = 75.48%
Currently selected inputs: g_wei, education, g_bmi, g_hei, dm_fg => Recog. rate = 77.5%
Selecting input 6:
Model 51/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, age} => Recog. rate = 77.98%
Model 52/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, occupation} => Recog. rate = 76.34%
Model 53/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, g_wc} => Recog. rate = 77.46%
Model 54/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, l_tg} => Recog. rate = 78.13%
Model 55/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, l_hdlc} => Recog. rate = 78.06%
Model 56/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, systolic} => Recog. rate = 77.46%
Model 57/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic} => Recog. rate = 78.20%
Currently selected inputs: g_wei, education, g_bmi, g_hei, dm_fg, diastolic => Recog. rate = 78.2%
Selecting input 7:
Model 58/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, age} => Recog. rate = 78.58%
Model 59/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, occupation} => Recog. rate = 77.94%
Model 60/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, g_wc} => Recog. rate = 77.87%
Model 61/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg} => Recog. rate = 79.62%
Model 62/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_hdlc} => Recog. rate = 79.06%
Model 63/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, systolic} => Recog. rate = 77.20%
Currently selected inputs: g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg => Recog. rate = 79.6%
Selecting input 8:
Model 64/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, age} => Recog. rate = 78.46%
Model 65/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, occupation} => Recog. rate = 78.02%
Model 66/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, g_wc} => Recog. rate = 78.73%
Model 67/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc} => Recog. rate = 79.14%
Model 68/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, systolic} => Recog. rate = 78.95%
Currently selected inputs: g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc => Recog. rate = 79.1%
Selecting input 9:
Model 69/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, age} => Recog. rate = 79.32%
Model 70/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, occupation} => Recog. rate = 78.28%
Model 71/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, g_wc} => Recog. rate = 79.36%
Model 72/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, systolic} => Recog. rate = 79.88%
Currently selected inputs: g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, systolic => Recog. rate = 79.9%
Selecting input 10:
Model 73/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, systolic, age} => Recog. rate = 80.10%
Model 74/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, systolic, occupation} => Recog. rate = 78.80%
Model 75/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, systolic, g_wc} => Recog. rate = 80.03%
Currently selected inputs: g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, systolic, age => Recog. rate = 80.1%
Selecting input 11:
Model 76/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, systolic, age, occupation} => Recog. rate = 77.65%
Model 77/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, systolic, age, g_wc} => Recog. rate = 79.69%
Currently selected inputs: g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, systolic, age, g_wc => Recog. rate = 79.7%
Selecting input 12:
Model 78/78: selected={g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, systolic, age, g_wc, occupation} => Recog. rate = 78.46%
Currently selected inputs: g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, systolic, age, g_wc, occupation => Recog. rate = 78.5%
Overall maximal recognition rate = 80.1%.
Selected 10 inputs (out of 12): g_wei, education, g_bmi, g_hei, dm_fg, diastolic, l_tg, l_hdlc, systolic, age
Elapsed time is 7.278278 seconds.
Dimensionality reduction based on LDA:
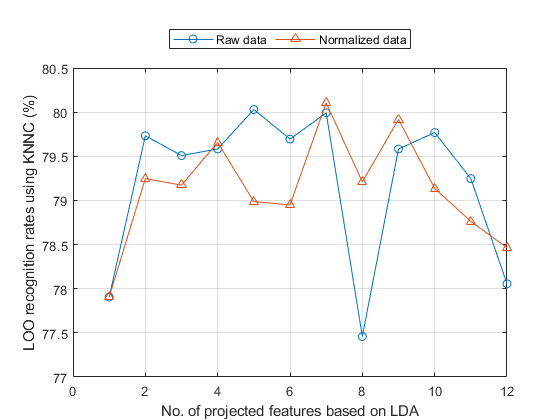
opt=ldaPerfViaKnncLoo('defaultOpt'); opt.mode='exact'; % This option causes error, why? tic recogRate1=ldaPerfViaKnncLoo(ds, opt); ds2=ds; ds2.input=inputNormalize(ds2.input); % input normalization recogRate2=ldaPerfViaKnncLoo(ds2, opt); fprintf('Time=%g sec\n', toc); [featureNum, dataNum] = size(ds.input); figure; plot(1:featureNum, 100*recogRate1, 'o-', 1:featureNum, 100*recogRate2, '^-'); grid on legend({'Raw data', 'Normalized data'}, 'location', 'northOutside', 'orientation', 'horizontal'); xlabel('No. of projected features based on LDA'); ylabel('LOO recognition rates using KNNC (%)');
Time=115.118 sec
Let's perform AutoML with ds:
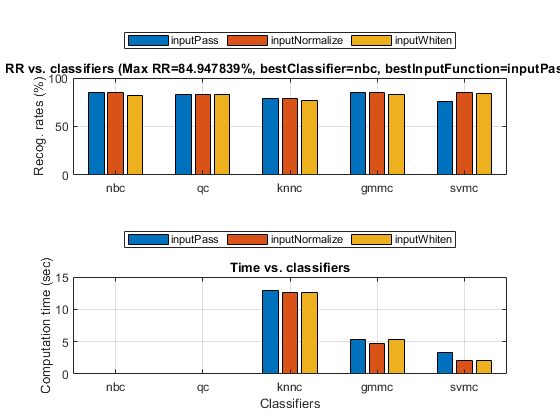
opt=perfCv4classifier('defaultOpt'); opt.foldNum=5; opt.classifiers(find(strcmp(opt.classifiers, 'src')))=[]; % Get rid of 'src' since it's extremely slow tic; [perfData, bestId]=perfCv4classifier(ds, opt, 1); toc %structDispInHtml(perfData, 'Performance of various classifiers via cross validation');
Elapsed time is 61.638805 seconds.
Plot of confusion matrix
confMat=confMatGet(ds.output, perfData(bestId).bestComputedClass);
opt=confMatPlot('defaultOpt');
opt.className=ds.outputName;
figure; confMatPlot(confMat, opt);
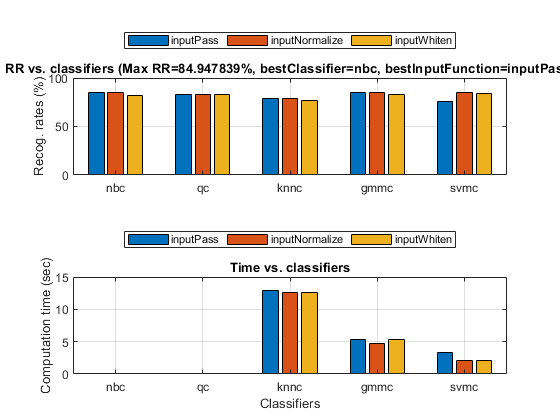
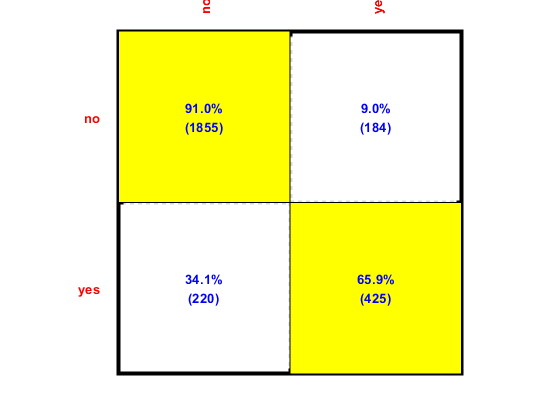
Since NBC is the best classifier, we'll use it for more detailed analysis. We can plot the DET curve:
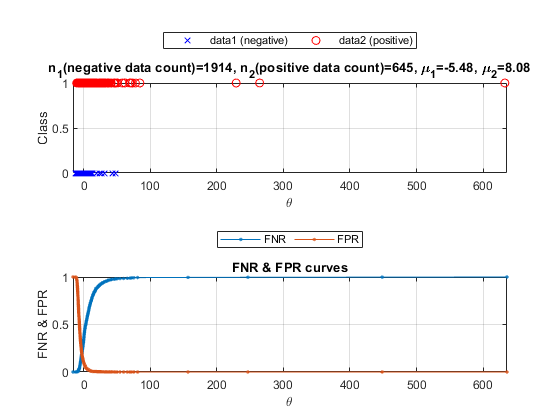
[cPrm, logLike1, rr1]=nbcTrain(ds); [computedClass, logLike2, rr2, hitIndex]=nbcEval(dsSmallTest, cPrm); logLike=diff(logLike2); id01=find(dsSmallTest.output==1); id02=find(dsSmallTest.output==2); data01=logLike(id01); data02=logLike(id02); opt=detPlot('defaultOpt'); opt.scaleMode='linear'; figure; detPlot(data01, data02, opt);
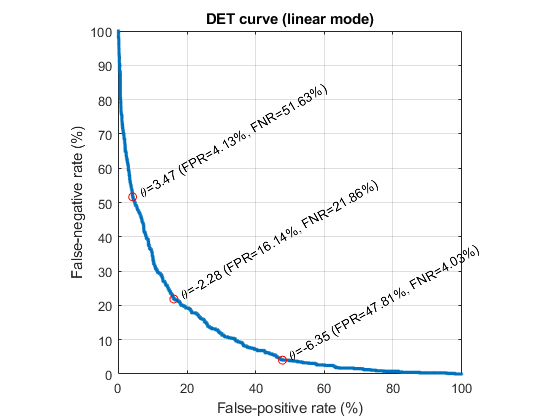
We can plot the density curves for each class:
opt=detDataGet('defaultOpt');
figure; [th, fp, fn]=detDataGet(data01, data02, opt, 1);
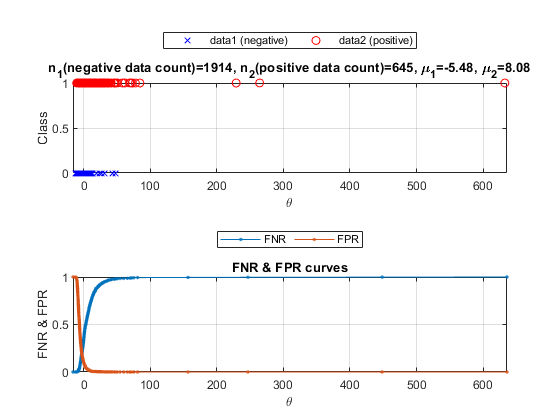
We can also plot the lift chart:
gt=dsSmallTest.output; opt=liftChart('defaultOpt'); figure; opt.type='binwisePrecision'; subplot(121); liftChart(logLike, gt, opt, 1); opt.type='cumulatedRecall'; subplot(122); liftChart(logLike, gt, opt, 1); figEnlarge
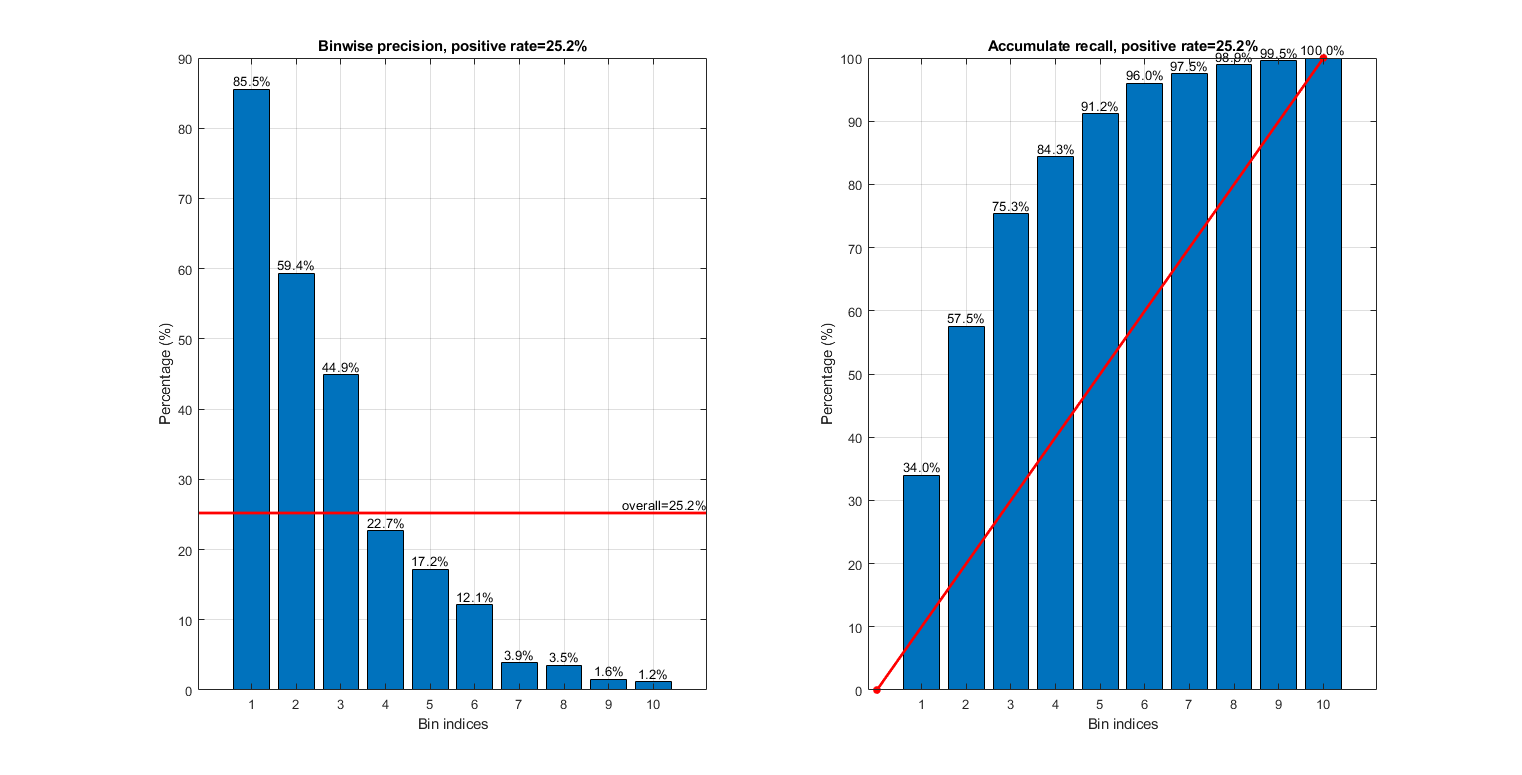
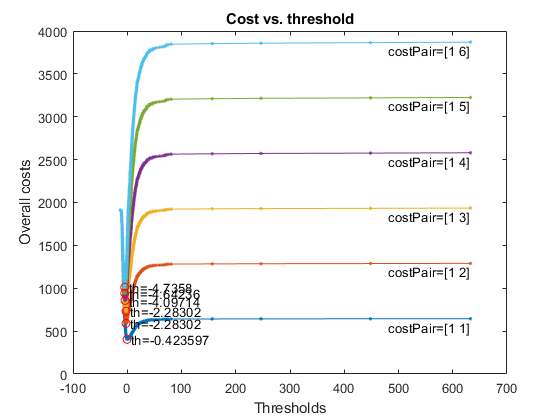
Since the selection of the best threshold depends on the cost of FP (false positive) and FN (false negative), we shall fix the cost of FP to be 1, and change the cost of FN to see the the best threshold varies, as follows:
figure;
opt=costVsTh4binClassifier('defaultOpt');
opt.fnCost=1:6;
[costMin, thBest, costVec, thVec, misclassifiedCount]=costVsTh4binClassifier(gt, logLike, opt, 1);
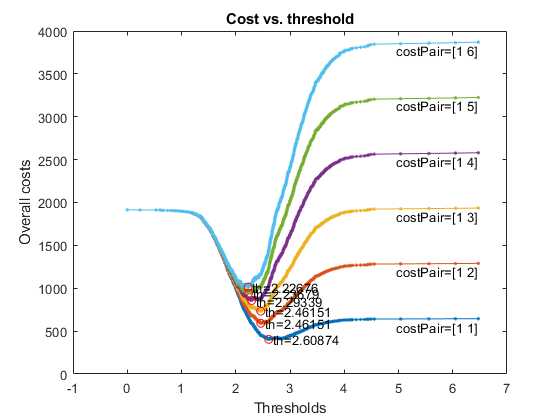
For better visualization, we can take the log of the likelihood:
figure; [costMin, thBest, costVec, thVec, misclassifiedCount]=costVsTh4binClassifier(gt, log(logLike-min(logLike)+1), opt, 1);
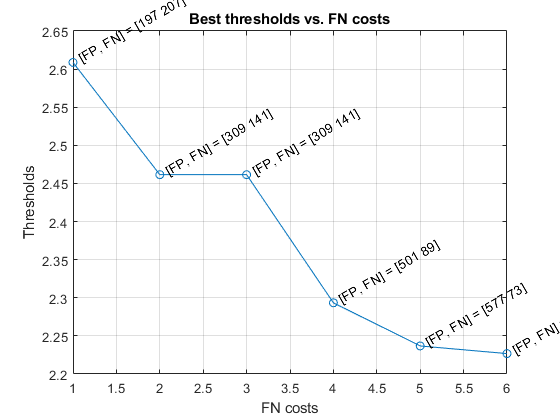
As best threshold vs. the cost of FN is shown next:
figure; plot(opt.fnCost, thBest, 'o-'); for i=1:length(opt.fnCost) h=text(opt.fnCost(i), thBest(i), sprintf(' [FP, FN] = %s', mat2str(misclassifiedCount{i}))); set(h, 'rotation', 30); end xlabel('FN costs'); ylabel('Thresholds'); title('Best thresholds vs. FN costs'); grid on
Since NBC is the best classifier, let's use it for feature selection:
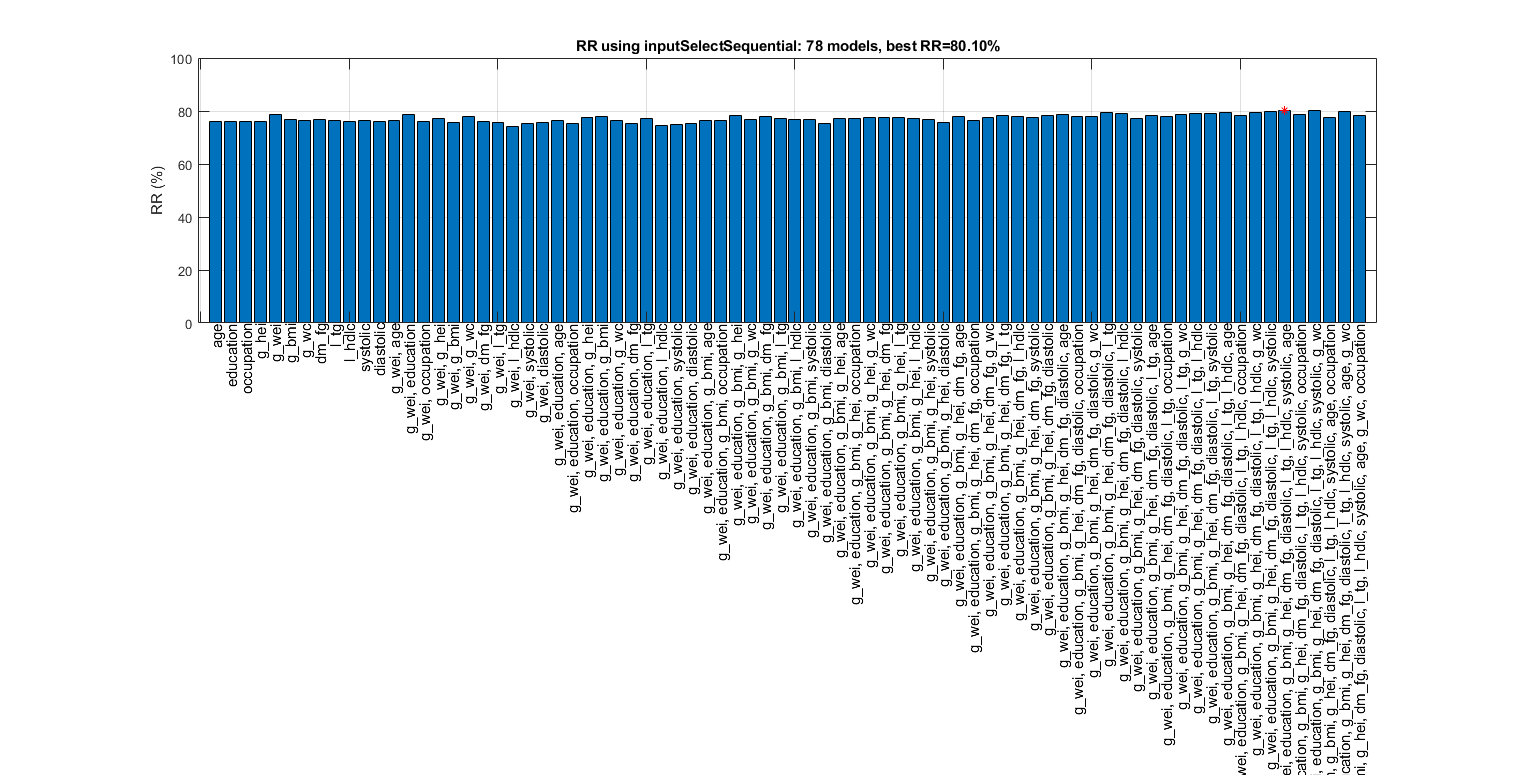
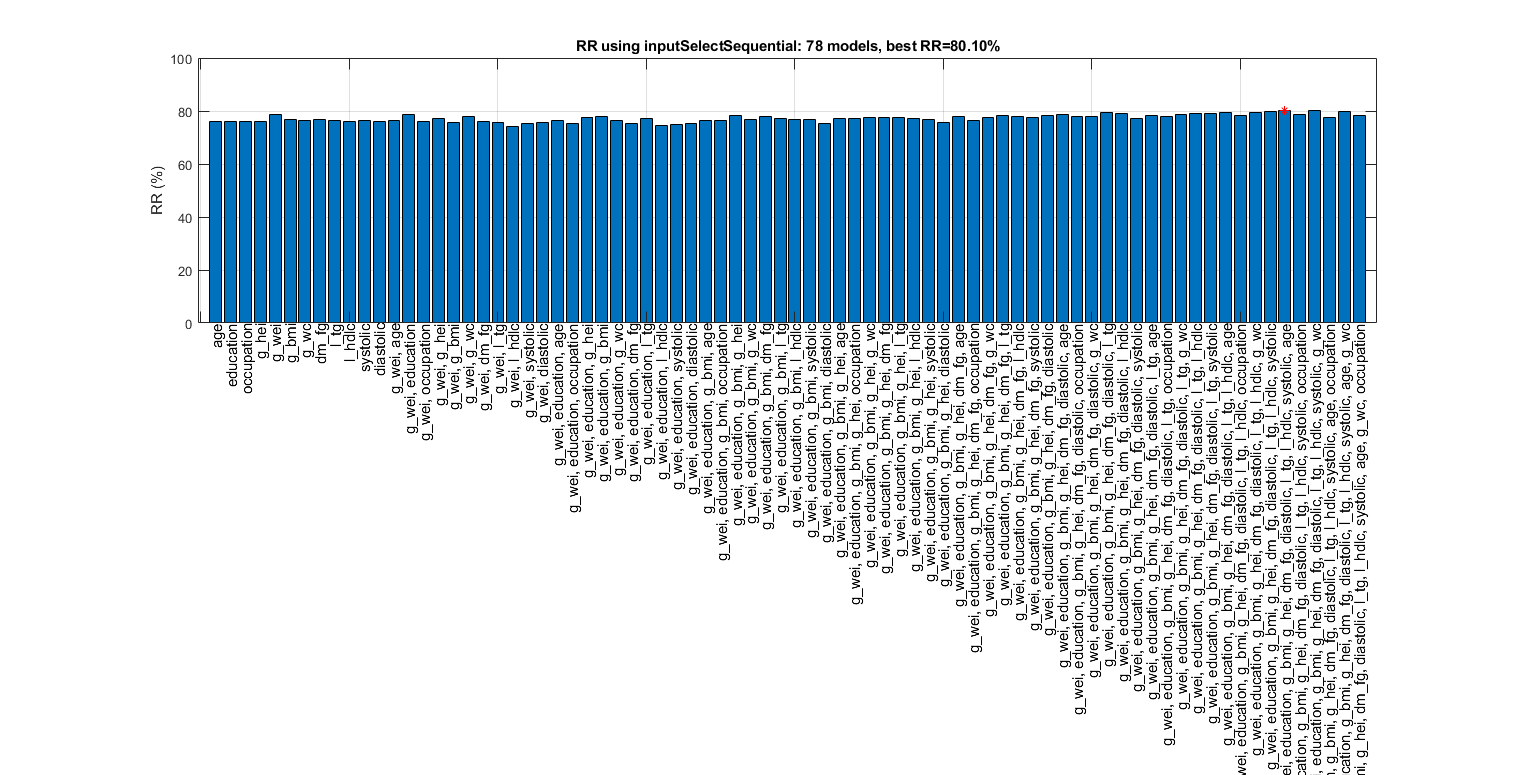
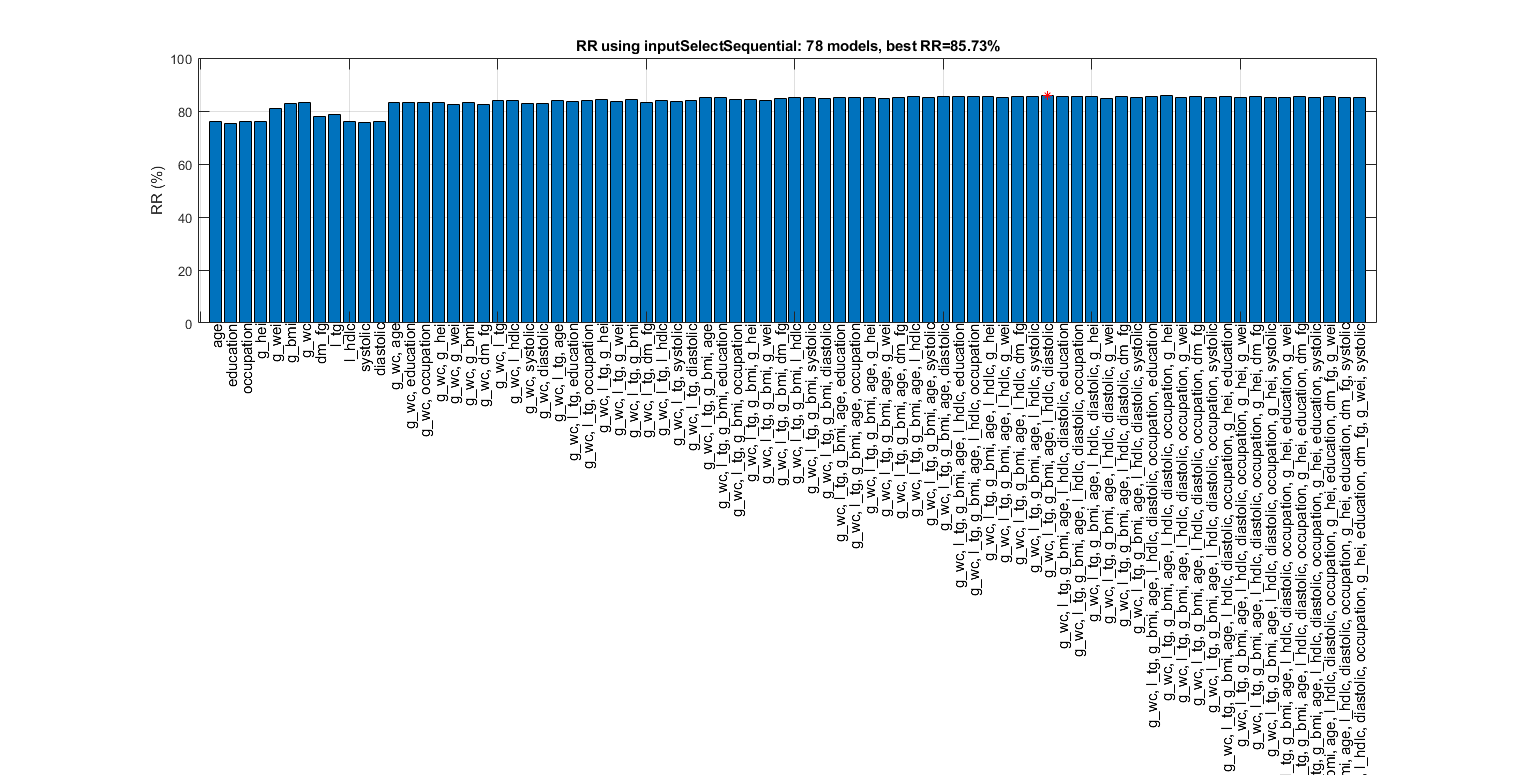
figure; tic; [inputId, bestRr]=inputSelectSequential(ds, inf, 'nbc'); toc; figEnlarge
Construct 78 "nbc" models, each with up to 12 inputs selected from all 12 inputs...
Selecting input 1:
Model 1/78: selected={age} => Recog. rate = 75.86%
Model 2/78: selected={education} => Recog. rate = 75.45%
Model 3/78: selected={occupation} => Recog. rate = 75.97%
Model 4/78: selected={g_hei} => Recog. rate = 75.97%
Model 5/78: selected={g_wei} => Recog. rate = 81.00%
Model 6/78: selected={g_bmi} => Recog. rate = 82.75%
Model 7/78: selected={g_wc} => Recog. rate = 83.05%
Model 8/78: selected={dm_fg} => Recog. rate = 78.09%
Model 9/78: selected={l_tg} => Recog. rate = 78.80%
Model 10/78: selected={l_hdlc} => Recog. rate = 76.01%
Model 11/78: selected={systolic} => Recog. rate = 75.82%
Model 12/78: selected={diastolic} => Recog. rate = 75.86%
Currently selected inputs: g_wc => Recog. rate = 83.0%
Selecting input 2:
Model 13/78: selected={g_wc, age} => Recog. rate = 83.27%
Model 14/78: selected={g_wc, education} => Recog. rate = 83.31%
Model 15/78: selected={g_wc, occupation} => Recog. rate = 83.38%
Model 16/78: selected={g_wc, g_hei} => Recog. rate = 83.12%
Model 17/78: selected={g_wc, g_wei} => Recog. rate = 82.64%
Model 18/78: selected={g_wc, g_bmi} => Recog. rate = 83.38%
Model 19/78: selected={g_wc, dm_fg} => Recog. rate = 82.34%
Model 20/78: selected={g_wc, l_tg} => Recog. rate = 83.98%
Model 21/78: selected={g_wc, l_hdlc} => Recog. rate = 83.98%
Model 22/78: selected={g_wc, systolic} => Recog. rate = 83.01%
Model 23/78: selected={g_wc, diastolic} => Recog. rate = 82.86%
Currently selected inputs: g_wc, l_tg => Recog. rate = 84.0%
Selecting input 3:
Model 24/78: selected={g_wc, l_tg, age} => Recog. rate = 83.90%
Model 25/78: selected={g_wc, l_tg, education} => Recog. rate = 83.46%
Model 26/78: selected={g_wc, l_tg, occupation} => Recog. rate = 83.90%
Model 27/78: selected={g_wc, l_tg, g_hei} => Recog. rate = 84.17%
Model 28/78: selected={g_wc, l_tg, g_wei} => Recog. rate = 83.72%
Model 29/78: selected={g_wc, l_tg, g_bmi} => Recog. rate = 84.39%
Model 30/78: selected={g_wc, l_tg, dm_fg} => Recog. rate = 83.16%
Model 31/78: selected={g_wc, l_tg, l_hdlc} => Recog. rate = 83.79%
Model 32/78: selected={g_wc, l_tg, systolic} => Recog. rate = 83.76%
Model 33/78: selected={g_wc, l_tg, diastolic} => Recog. rate = 84.05%
Currently selected inputs: g_wc, l_tg, g_bmi => Recog. rate = 84.4%
Selecting input 4:
Model 34/78: selected={g_wc, l_tg, g_bmi, age} => Recog. rate = 85.13%
Model 35/78: selected={g_wc, l_tg, g_bmi, education} => Recog. rate = 84.95%
Model 36/78: selected={g_wc, l_tg, g_bmi, occupation} => Recog. rate = 84.46%
Model 37/78: selected={g_wc, l_tg, g_bmi, g_hei} => Recog. rate = 84.46%
Model 38/78: selected={g_wc, l_tg, g_bmi, g_wei} => Recog. rate = 83.87%
Model 39/78: selected={g_wc, l_tg, g_bmi, dm_fg} => Recog. rate = 84.80%
Model 40/78: selected={g_wc, l_tg, g_bmi, l_hdlc} => Recog. rate = 84.99%
Model 41/78: selected={g_wc, l_tg, g_bmi, systolic} => Recog. rate = 84.95%
Model 42/78: selected={g_wc, l_tg, g_bmi, diastolic} => Recog. rate = 84.80%
Currently selected inputs: g_wc, l_tg, g_bmi, age => Recog. rate = 85.1%
Selecting input 5:
Model 43/78: selected={g_wc, l_tg, g_bmi, age, education} => Recog. rate = 85.13%
Model 44/78: selected={g_wc, l_tg, g_bmi, age, occupation} => Recog. rate = 84.99%
Model 45/78: selected={g_wc, l_tg, g_bmi, age, g_hei} => Recog. rate = 85.25%
Model 46/78: selected={g_wc, l_tg, g_bmi, age, g_wei} => Recog. rate = 84.72%
Model 47/78: selected={g_wc, l_tg, g_bmi, age, dm_fg} => Recog. rate = 85.17%
Model 48/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc} => Recog. rate = 85.58%
Model 49/78: selected={g_wc, l_tg, g_bmi, age, systolic} => Recog. rate = 85.17%
Model 50/78: selected={g_wc, l_tg, g_bmi, age, diastolic} => Recog. rate = 85.36%
Currently selected inputs: g_wc, l_tg, g_bmi, age, l_hdlc => Recog. rate = 85.6%
Selecting input 6:
Model 51/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, education} => Recog. rate = 85.47%
Model 52/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, occupation} => Recog. rate = 85.58%
Model 53/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, g_hei} => Recog. rate = 85.51%
Model 54/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, g_wei} => Recog. rate = 85.13%
Model 55/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, dm_fg} => Recog. rate = 85.47%
Model 56/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, systolic} => Recog. rate = 85.54%
Model 57/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic} => Recog. rate = 85.73%
Currently selected inputs: g_wc, l_tg, g_bmi, age, l_hdlc, diastolic => Recog. rate = 85.7%
Selecting input 7:
Model 58/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, education} => Recog. rate = 85.43%
Model 59/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation} => Recog. rate = 85.66%
Model 60/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, g_hei} => Recog. rate = 85.47%
Model 61/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, g_wei} => Recog. rate = 84.80%
Model 62/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, dm_fg} => Recog. rate = 85.32%
Model 63/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, systolic} => Recog. rate = 85.17%
Currently selected inputs: g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation => Recog. rate = 85.7%
Selecting input 8:
Model 64/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, education} => Recog. rate = 85.39%
Model 65/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei} => Recog. rate = 85.69%
Model 66/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_wei} => Recog. rate = 85.02%
Model 67/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, dm_fg} => Recog. rate = 85.43%
Model 68/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, systolic} => Recog. rate = 85.13%
Currently selected inputs: g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei => Recog. rate = 85.7%
Selecting input 9:
Model 69/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, education} => Recog. rate = 85.39%
Model 70/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, g_wei} => Recog. rate = 85.21%
Model 71/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, dm_fg} => Recog. rate = 85.28%
Model 72/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, systolic} => Recog. rate = 85.21%
Currently selected inputs: g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, education => Recog. rate = 85.4%
Selecting input 10:
Model 73/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, education, g_wei} => Recog. rate = 85.13%
Model 74/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, education, dm_fg} => Recog. rate = 85.28%
Model 75/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, education, systolic} => Recog. rate = 84.99%
Currently selected inputs: g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, education, dm_fg => Recog. rate = 85.3%
Selecting input 11:
Model 76/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, education, dm_fg, g_wei} => Recog. rate = 85.47%
Model 77/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, education, dm_fg, systolic} => Recog. rate = 85.06%
Currently selected inputs: g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, education, dm_fg, g_wei => Recog. rate = 85.5%
Selecting input 12:
Model 78/78: selected={g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, education, dm_fg, g_wei, systolic} => Recog. rate = 85.17%
Currently selected inputs: g_wc, l_tg, g_bmi, age, l_hdlc, diastolic, occupation, g_hei, education, dm_fg, g_wei, systolic => Recog. rate = 85.2%
Overall maximal recognition rate = 85.7%.
Selected 6 inputs (out of 12): g_wc, l_tg, g_bmi, age, l_hdlc, diastolic
Elapsed time is 420.385532 seconds.
Now we can list the training and test performance of the dataset based on NBC, with breakdowns into genders and years:
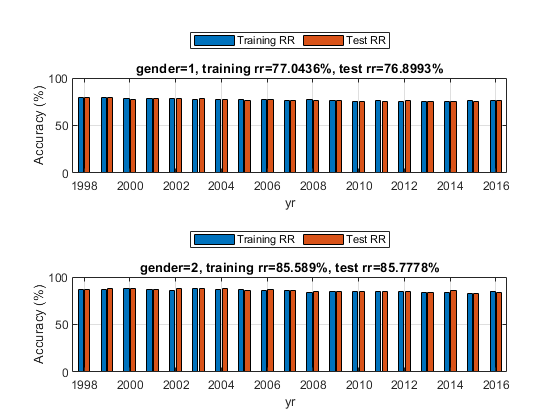
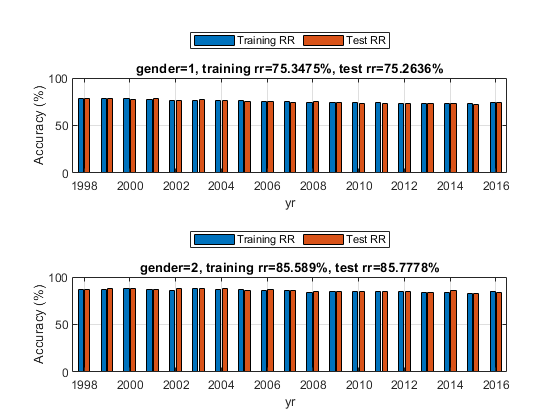
figure; fprintf('Holdout test on female data:\n'); opt1=struct('inputName', 'gender', 'inputValue', 1); dsFemaleTrain=dsSubset(dsTrain, opt1); dsFemaleTest= dsSubset(dsTest, opt1); optField=perfByField('defaultOpt'); optField.fieldName='yr'; optField.classifier='nbc'; subplot(211); rrFemale=perfByField(dsFemaleTrain, dsFemaleTest, optField, 1); title(sprintf('gender=%d, training rr=%g%%, test rr=%g%%', 1, 100*mean(rrFemale(:,1)), 100*mean(rrFemale(:,2)))); fprintf('Holdout test on male data:\n'); opt2=struct('inputName', 'gender', 'inputValue', 2); dsMaleTrain=dsSubset(dsTrain, opt2); dsMaleTest= dsSubset(dsTest, opt2); subplot(212); rrMale=perfByField(dsMaleTrain, dsMaleTest, optField, 1); title(sprintf('gender=%d, training rr=%g%%, test rr=%g%%', 2, 100*mean(rrMale(:,1)), 100*mean(rrMale(:,2))));
Holdout test on female data: 1/19: yr=1998, training rr=79.0122%, test rr=79.0402% 2/19: yr=1999, training rr=79.444%, test rr=79.3834% 3/19: yr=2000, training rr=78.6279%, test rr=77.3954% 4/19: yr=2001, training rr=78.6651%, test rr=78.1515% 5/19: yr=2002, training rr=77.9463%, test rr=78.277% 6/19: yr=2003, training rr=77.3847%, test rr=78.267% 7/19: yr=2004, training rr=77.6685%, test rr=77.4722% 8/19: yr=2005, training rr=77.7899%, test rr=76.5361% 9/19: yr=2006, training rr=77.0638%, test rr=76.9408% 10/19: yr=2007, training rr=76.4965%, test rr=76.8007% 11/19: yr=2008, training rr=76.9514%, test rr=76.2332% 12/19: yr=2009, training rr=76.4996%, test rr=76.3547% 13/19: yr=2010, training rr=75.7544%, test rr=75.6582% 14/19: yr=2011, training rr=76.4349%, test rr=75.6515% 15/19: yr=2012, training rr=75.3544%, test rr=76.1861% 16/19: yr=2013, training rr=75.2408%, test rr=74.875% 17/19: yr=2014, training rr=75.4434%, test rr=75.608% 18/19: yr=2015, training rr=75.8489%, test rr=75.5326% 19/19: yr=2016, training rr=76.2015%, test rr=76.7237% Holdout test on male data: 1/19: yr=1998, training rr=86.4635%, test rr=86.7281% 2/19: yr=1999, training rr=86.9862%, test rr=87.6339% 3/19: yr=2000, training rr=87.9987%, test rr=87.5893% 4/19: yr=2001, training rr=86.7384%, test rr=87.1166% 5/19: yr=2002, training rr=85.9074%, test rr=87.3848% 6/19: yr=2003, training rr=87.5606%, test rr=87.9708% 7/19: yr=2004, training rr=87.308%, test rr=87.474% 8/19: yr=2005, training rr=86.8787%, test rr=86.2822% 9/19: yr=2006, training rr=85.816%, test rr=87.0005% 10/19: yr=2007, training rr=86.2485%, test rr=86.0534% 11/19: yr=2008, training rr=84.1669%, test rr=84.2796% 12/19: yr=2009, training rr=84.9519%, test rr=84.3178% 13/19: yr=2010, training rr=84.7205%, test rr=84.8768% 14/19: yr=2011, training rr=84.2789%, test rr=85.0524% 15/19: yr=2012, training rr=84.4606%, test rr=84.5538% 16/19: yr=2013, training rr=83.5204%, test rr=83.6188% 17/19: yr=2014, training rr=84.1688%, test rr=85.3086% 18/19: yr=2015, training rr=82.8082%, test rr=82.7931% 19/19: yr=2016, training rr=85.2086%, test rr=83.7436%
We can only use the selected inputs for the above plot:
figure; fprintf('Holdout test on female data:\n'); opt1=struct('inputName', 'gender', 'inputValue', 1); dsFemaleTrain=dsSubset(dsTrain, opt1); dsFemaleTrain.input=dsFemaleTrain.input(inputId,:); dsFemaleTrain.inputName=dsFemaleTrain.inputName(inputId); dsFemaleTest= dsSubset(dsTest, opt1); dsFemaleTest.input= dsFemaleTest.input(inputId,:); dsFemaleTest.inputName= dsFemaleTest.inputName(inputId); subplot(211); rrFemale=perfByField(dsFemaleTrain, dsFemaleTest, optField, 1); title(sprintf('gender=%d, training rr=%g%%, test rr=%g%%', 1, 100*mean(rrFemale(:,1)), 100*mean(rrFemale(:,2)))); fprintf('Holdout test on male data:\n'); opt2=struct('inputName', 'gender', 'inputValue', 2); dsMaleTrain=dsSubset(dsTrain, opt2); dsMaleTest= dsSubset(dsTest, opt2); subplot(212); rrMale=perfByField(dsMaleTrain, dsMaleTest, optField, 1); title(sprintf('gender=%d, training rr=%g%%, test rr=%g%%', 2, 100*mean(rrMale(:,1)), 100*mean(rrMale(:,2))));
Holdout test on female data: 1/19: yr=1998, training rr=78.5079%, test rr=78.6875% 2/19: yr=1999, training rr=78.2526%, test rr=78.6982% 3/19: yr=2000, training rr=78.8105%, test rr=77.6683% 4/19: yr=2001, training rr=77.8018%, test rr=78.035% 5/19: yr=2002, training rr=76.3349%, test rr=76.8206% 6/19: yr=2003, training rr=75.9064%, test rr=77.2455% 7/19: yr=2004, training rr=76.2328%, test rr=75.9777% 8/19: yr=2005, training rr=76.0754%, test rr=75.1334% 9/19: yr=2006, training rr=75.4752%, test rr=75.7742% 10/19: yr=2007, training rr=75.1755%, test rr=74.4958% 11/19: yr=2008, training rr=74.513%, test rr=74.8879% 12/19: yr=2009, training rr=74.2109%, test rr=74.4439% 13/19: yr=2010, training rr=73.7825%, test rr=73.6723% 14/19: yr=2011, training rr=73.8769%, test rr=73.4528% 15/19: yr=2012, training rr=72.8462%, test rr=73.0079% 16/19: yr=2013, training rr=73.1716%, test rr=72.6964% 17/19: yr=2014, training rr=72.8172%, test rr=73.2111% 18/19: yr=2015, training rr=73.4581%, test rr=72.1993% 19/19: yr=2016, training rr=74.353%, test rr=73.901% Holdout test on male data: 1/19: yr=1998, training rr=86.4635%, test rr=86.7281% 2/19: yr=1999, training rr=86.9862%, test rr=87.6339% 3/19: yr=2000, training rr=87.9987%, test rr=87.5893% 4/19: yr=2001, training rr=86.7384%, test rr=87.1166% 5/19: yr=2002, training rr=85.9074%, test rr=87.3848% 6/19: yr=2003, training rr=87.5606%, test rr=87.9708% 7/19: yr=2004, training rr=87.308%, test rr=87.474% 8/19: yr=2005, training rr=86.8787%, test rr=86.2822% 9/19: yr=2006, training rr=85.816%, test rr=87.0005% 10/19: yr=2007, training rr=86.2485%, test rr=86.0534% 11/19: yr=2008, training rr=84.1669%, test rr=84.2796% 12/19: yr=2009, training rr=84.9519%, test rr=84.3178% 13/19: yr=2010, training rr=84.7205%, test rr=84.8768% 14/19: yr=2011, training rr=84.2789%, test rr=85.0524% 15/19: yr=2012, training rr=84.4606%, test rr=84.5538% 16/19: yr=2013, training rr=83.5204%, test rr=83.6188% 17/19: yr=2014, training rr=84.1688%, test rr=85.3086% 18/19: yr=2015, training rr=82.8082%, test rr=82.7931% 19/19: yr=2016, training rr=85.2086%, test rr=83.7436%
Summary
This is a brief tutorial on fatty liver recognition based on patients' data. There are several directions for further improvement:
- Try the classification problem using the whole dataset
- Use template matching as an alternative to improve the performance
Appendix
List of functions, scripts, and datasets used in this script:
Date and time when finishing this script:
fprintf('Date & time: %s\n', char(datetime));
Date & time: 24-Apr-2022 23:39:01
Overall elapsed time:
toc(scriptStartTime)
Elapsed time is 673.673540 seconds.
Jyh-Shing Roger Jang, created on
datetime
ans = datetime 24-Apr-2022 23:39:01